Difference between revisions of "HackteriaLab 2013 Commons"
(→Fluorescent Reporter Bacteria for Arsenic Sensing) |
|||
| (20 intermediate revisions by 3 users not shown) | |||
| Line 4: | Line 4: | ||
Start putting down ideas... | Start putting down ideas... | ||
| − | == Fluorescent Reporter Bacteria for Arsenic Sensing == | + | == Fluorescent Reporter Bacteria for Arsenic Sensing == |
| + | (still editing, not completed) | ||
| + | <br><br> | ||
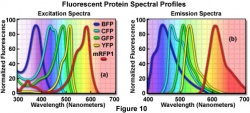
| + | This Arsenic sensing [http://en.wikipedia.org/wiki/Green_fluorescent_protein GFP] [http://en.wikipedia.org/wiki/Reporter_gene reporter bacteria] was made in the laboratory of [http://www.unil.ch/dmf/page15250_en.html Professor Jan van der Meer] at the University of Lausanne. | ||
| − | ... | + | The arsenic reporter bacteria is built on what makes many naturally occuring bacteria resistant (i.e. won't die in the presence of) arsenite and arsenate. |
| + | <br><br> | ||
| + | [[File:arsenic-repressed.jpg | 300px]] | ||
| + | <br><br> | ||
| + | Sensor element is the ArsR (in red), which is a protein that has 2 functions:<br> | ||
| + | - binds to a specific region of DNA (bright green boxes on the linear representation of DNA)<br> | ||
| + | - to bind to arsenite <br> | ||
| + | |||
| + | Sensor output is the GFP (green fluorescent protein) expression. The gene for GFP (represented by the bright green arrow on the DNA) is engineered to be behind the repressive element.<br> | ||
| + | The dark green arrow is the ArsR gene. In other words, in this design, more aresenite the bacteria "sees", the more repressor element it expresses.<br><br> | ||
| + | <br><br> | ||
| + | [[File:arsenic-reporting.jpg | 300px]] | ||
| + | <br><br> | ||
| + | In the presence of arsenite (As), which enters the bacteria passively, the ArsR prefers to bind to As rather than the repressive element. This makes room for [http://en.wikipedia.org/wiki/Transcription_%28genetics%29 gene transcription] to start. Transcription, Translation into GFP protein. Under UV light, or at ~400 or ~488nm excitation, the protein fluorescence can be measured or visualized. <br><br><br> | ||
| + | |||
| + | The details can be found in this manuscript: Stocker, J., D. Balluch, et al. (2003). Environ. Sci. Technol. 37: 4743-4750. | ||
| + | |||
| + | The reporter bacteria is in the common laboratory ''E. coli'' DH5α strain used in molecular biology. They can be easily grown using standard molecular biology techniques in LB, with kanamycin as the antibiotic used for selecting the bacteria with the arsenic GFP reporter.<br> | ||
| + | |||
| + | This bacteria was made available to us as a part of an international collaboration [http://biodesign.cc BIO-DESIGN for the REAL WORLD] between [http://artscienceblr.org/ (Art)ScienceBLR], [http://lifepatch.org Lifepatch], and [http://wiki.epfl.ch/biodesign-realworld EPFL] to design a sensor for arsenic and/or ''E. coli'' in the local waters. The bacteria was shipped to the [http://www.ncbs.res.in/ NCBS]. | ||
| + | <br><br> | ||
== Fluorescent Microscopy == | == Fluorescent Microscopy == | ||
| Line 18: | Line 41: | ||
<br> | <br> | ||
UV LEDs:<br> | UV LEDs:<br> | ||
| − | http://www.ebay.com/itm/10-3mm-2-Pin-UV-Purple-LED-Light-Lamp-Bright-5000-Mcd-/150961264600?pt=LH_DefaultDomain_0&hash=item2325fe17d8 <br> | + | http://www.ebay.com/itm/10-3mm-2-Pin-UV-Purple-LED-Light-Lamp-Bright-5000-Mcd-/150961264600?pt=LH_DefaultDomain_0&hash=item2325fe17d8 <br><br> |
| + | |||
| + | [http://www.plosone.org/article/info%3Adoi%2F10.1371%2Fjournal.pone.0011890 Portable, Battery-Operated, Low-Cost, Bright Field and Fluorescence Microscope], Miller et al. PlosOne <br><br> | ||
== Building Bio Domes == | == Building Bio Domes == | ||
| Line 60: | Line 85: | ||
http://www.wired.com/design/2013/01/diy-bio-printer/ | http://www.wired.com/design/2013/01/diy-bio-printer/ | ||
| + | |||
| + | ... done that at last HackteriaLab! see [[DIY Micro Dispensing and Bio Printing]] | ||
| + | |||
| + | ===Poor Man's BioPrinter=== | ||
| + | |||
| + | ==Rice Beer== | ||
| + | |||
| + | Making Chyaang / Tho: A Traditional Rice Beer From The Kathmandu Valley | ||
| + | |||
| + | http://www.karkhana.asia/making-chyaang-tho/ | ||
| + | |||
| + | ==The Making of an Euglena Burger== | ||
| + | |||
| + | [[File:euglena_culture_7days_light.jpg|250px]] | ||
| + | |||
| + | Mmmmmhhhh, half animal, half plant! | ||
| + | |||
| + | |||
| + | ==Hydra Hacks== | ||
| + | |||
| + | [[File:hydraHacks.jpg|320px]] | ||
| + | |||
| + | |||
| + | ==The Hunt for the Coloured MicroBugs== | ||
| + | |||
| + | [[File:Sakar_bug_hunting.jpg|320px]] | ||
| + | |||
| + | [[File:Sakar_dish_after2days.jpg|320px]] | ||
Latest revision as of 19:17, 5 March 2013
<- back to main page of HackteriaLab_2013 - Bangalore
Start putting down ideas...
Contents
Fluorescent Reporter Bacteria for Arsenic Sensing
(still editing, not completed)
This Arsenic sensing GFP reporter bacteria was made in the laboratory of Professor Jan van der Meer at the University of Lausanne.
The arsenic reporter bacteria is built on what makes many naturally occuring bacteria resistant (i.e. won't die in the presence of) arsenite and arsenate.
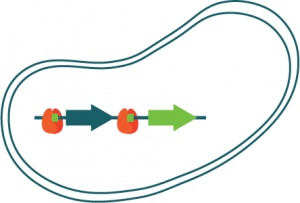
Sensor element is the ArsR (in red), which is a protein that has 2 functions:
- binds to a specific region of DNA (bright green boxes on the linear representation of DNA)
- to bind to arsenite
Sensor output is the GFP (green fluorescent protein) expression. The gene for GFP (represented by the bright green arrow on the DNA) is engineered to be behind the repressive element.
The dark green arrow is the ArsR gene. In other words, in this design, more aresenite the bacteria "sees", the more repressor element it expresses.
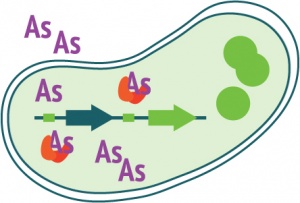
In the presence of arsenite (As), which enters the bacteria passively, the ArsR prefers to bind to As rather than the repressive element. This makes room for gene transcription to start. Transcription, Translation into GFP protein. Under UV light, or at ~400 or ~488nm excitation, the protein fluorescence can be measured or visualized.
The details can be found in this manuscript: Stocker, J., D. Balluch, et al. (2003). Environ. Sci. Technol. 37: 4743-4750.
The reporter bacteria is in the common laboratory E. coli DH5α strain used in molecular biology. They can be easily grown using standard molecular biology techniques in LB, with kanamycin as the antibiotic used for selecting the bacteria with the arsenic GFP reporter.
This bacteria was made available to us as a part of an international collaboration BIO-DESIGN for the REAL WORLD between (Art)ScienceBLR, Lifepatch, and EPFL to design a sensor for arsenic and/or E. coli in the local waters. The bacteria was shipped to the NCBS.
Fluorescent Microscopy
Build a fluorescence microscope based on the Hackteria webcam kit by adding UV-LEDs. Eventualy modifying the webcam to longer exposure to increase sensitivity. Inspect different samples to find fluorescent material. Modify samples (blood cells) to be fluorescent and try to sort them based in microscopy imaging.
source:microscopy resource center
WebCam long exposure hack:
http://www.pmdo.com/wintro.htm
UV LEDs:
http://www.ebay.com/itm/10-3mm-2-Pin-UV-Purple-LED-Light-Lamp-Bright-5000-Mcd-/150961264600?pt=LH_DefaultDomain_0&hash=item2325fe17d8
Portable, Battery-Operated, Low-Cost, Bright Field and Fluorescence Microscope, Miller et al. PlosOne
Building Bio Domes

from Yashas:
http://www.byexample.com/projects/current/dome_construction
http://2011.igem.org/Team:ArtScienceBangalore/Outreach
Grow real Computer Bugs
In 1947, Grace Murray Hopper was working Mark II Computer when the machine was experiencing problems. An investigation showed that there was a moth trapped between the points of Relay #70, in Panel F. This was the "First actual case of bug being found." The computer was debugged - and what happed to the bug that actual tried to "reprogram" the computer and make the first steps in bio-computing? It was affixed in the log book of Mr. Hopper. It was actually a moth and since it did not follow moore's law it was somehow forgotten. We want to identify and rediscover the famous animal. Find out what other bugs have been active in bugging (in fact the modern meaning can be dated back to at least 1896). Grow them and give them a new chance to be part of an "Art-Sci Bio Computer Project". For example a BioDuino micro controller board with a real bug actively reprogramming it...

Photo of the first recorded computer bug.
http://www.jamesshuggins.com/h/tek1/first_computer_bug.htm
Shruti Hacking
Take the shruti hacking started in 2012 to a new level. Approach Radel to get more devices (for free (and pay flights for some talented music artists to come to Bangalore)) and bring new spirit into these amazing boxes. Try to fully understand the previously reverse engineered multi chorous audio system. Spend long nights immersing into the sound of shruti.
Radel Electronics Pvt. Ltd. is the pioneer in the field of electronic musical instruments for Indian music. Radel started in a garage in 1979 and has today state-of-the-art facilities for designing and manufacturing a wide range of Digital Indian musical instruments.

http://wiki.sgmk-ssam.ch/index.php/Shruti_Hacking_-_Radel_Nano_Dx
Plantsensors
http://tangible.media.mit.edu/projects/plantsensor/
DIY BioPrinter
http://www.wired.com/design/2013/01/diy-bio-printer/
... done that at last HackteriaLab! see DIY Micro Dispensing and Bio Printing
Poor Man's BioPrinter
Rice Beer
Making Chyaang / Tho: A Traditional Rice Beer From The Kathmandu Valley
http://www.karkhana.asia/making-chyaang-tho/
The Making of an Euglena Burger
Mmmmmhhhh, half animal, half plant!
Hydra Hacks